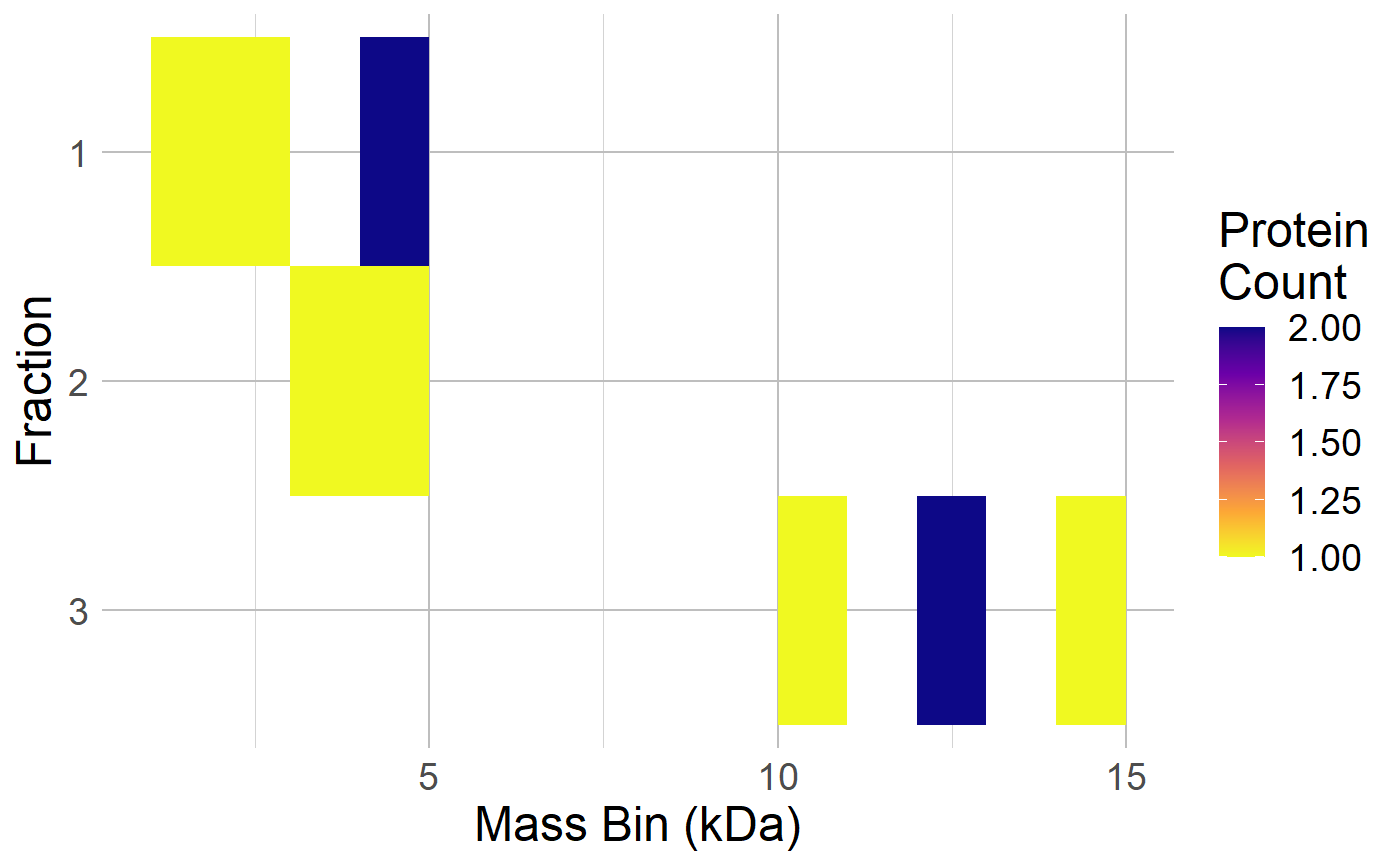
Uses a data frame containing mass and fraction columns to generate a heatmap showing the distribution of masses per fraction.
make_heatmap( df, plotType = "Protein", orientation = "h", binSize = 1000, savePDF = FALSE, outputDir = getwd(), outputPrefix = format(Sys.time(), "%Y%m%d_%H%M%S"), massColname = "mass", fractionColname = "fraction", axisRange = NULL, countRange = NULL )
Arguments
| df | A data frame containing columns for mass (in Daltons) and fraction. |
|---|---|
| plotType | Type of heatmap to create, typically "Protein" or "Proteoform".This only affects the legend title and output file name and can be any string. |
| orientation | Controls orientation of heatmap. Set to "h" for horizontal (mass bins on X-axis) or "v" for vertical. |
| binSize | Size of the mass bin in Daltons. Masses in the data frame should be in Daltons. |
| savePDF | Boolean value (TRUE or FALSE). Should a PDF be saved to the output directory? |
| outputDir | Directory to place output files into. |
| outputPrefix | String to use to prepend heatmap output filenames. Defaults to date and time. |
| massColname | Name of column containing masses. Defaults to "mass". |
| fractionColname | Name of column containing fractions. Defaults to "fraction". |
| axisRange | Numeric vector of length 2 specifying range of heatmap in kDa. Defaults to NULL, and range is determined by rounding highest mass bin up to the nearest interval of 5. |
| countRange | Numeric vector of length 2 specifying range of counts to use for generating heatmap fill color range. Defaults to NULL, which determines color range automatically using ggplot2::scale_fill_viridis_c(option = "C", direction = -1). Useful for creating multiple heatmaps with matching fill color ranges. |
Value
A heatmap (a ggplot object)
Examples
df <- data.frame( "fraction" = c(1,1,1,1,2,2,3,3,3,3), "mass" = c(1500,3000,4200,4250,3500,4500,10500,12050,12075,14050) ) make_heatmap( df, plotType = "Protein", savePDF = F, binSize = 1000 )